Getting Started¶
Importing MPDAF¶
MPDAF is divided into sub-packages, each of which is composed of several
classes. The following example shows how to import the Cube and
PixTable classes:
In [1]: from mpdaf.obj import Cube
In [2]: from mpdaf.drs import PixTable
All of the examples in the MPDAF web pages are shown being typed into
an interactive IPython shell. This shell is the origin of the prompts
like In [1]: in the above example. The examples can also be entered
in other shells, such as the native Python shell.
Loading your first MUSE datacube¶
MUSE datacubes are generally loaded from FITS files. In these files the fluxes and variances are stored in separate FITS extensions. For example:
# data and variance arrays are read from DATA and STAT extensions of the file
In [3]: cube = Cube('../data/obj/CUBE.fits')
In [4]: cube.info()
[INFO] 1595 x 10 x 20 Cube (../data/obj/CUBE.fits)
[INFO] .data(1595 x 10 x 20) (1e-40 erg / (Angstrom cm2 s)), .var(1595 x 10 x 20)
[INFO] center:(-30:00:00.4494,01:20:00.4376) size in arcsec:(2.000,4.000) step in arcsec:(0.200,0.200) rot:-0.0 deg
[INFO] wavelength: min:7300.00 max:9292.50 step:1.25 Angstrom
The listed dimensions of the cube, 1595 x 10 x 20, indicate that the cube has 1595 spectral pixels and 10 x 20 spatial pixels. The order in which these dimensions are listed, follows the indexing conventions used by Python to handle 3D arrays (see Spectrum, Image and Cube format for more information).
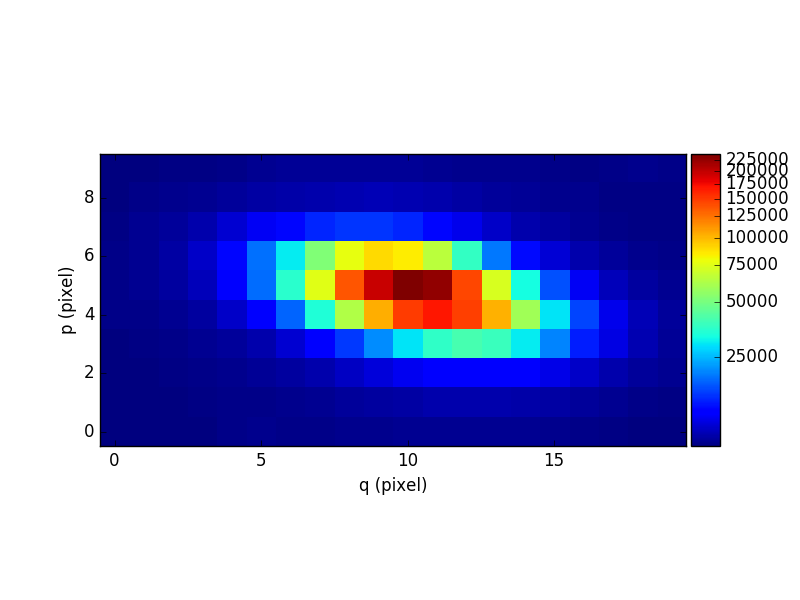
Let’s compute the reconstructed white-light image and display it. The white-light image is obtained by summing each spatial pixel of the cube along the wavelength axis. This converts the 3D cube into a 2D image.
In [5]: ima = cube.sum(axis=0)
In [6]: type(ima)
Out[6]: mpdaf.obj.image.Image
In [7]: plt.figure()
Out[7]: <matplotlib.figure.Figure at 0x7fb735f11350>
In [8]: ima.plot(scale='arcsinh', colorbar='v')
Out[8]: <matplotlib.image.AxesImage at 0x7fb74446f190>
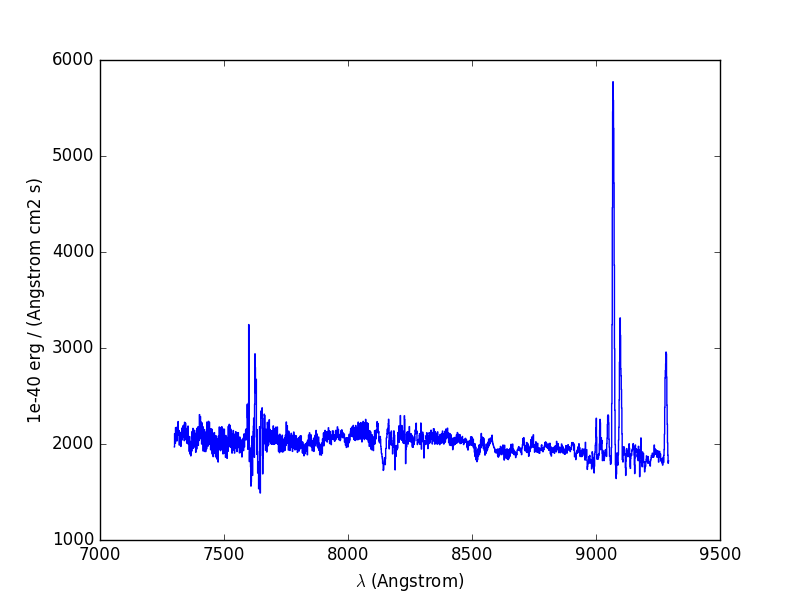
Let’s now compute the overall spectrum of the cube by taking the cube and summing along the X and Y axes of the image plane. This yields the total flux per spectral pixel.
In [9]: sp = cube.sum(axis=(1,2))
In [10]: type(sp)
Out[10]: mpdaf.obj.spectrum.Spectrum
In [11]: plt.figure()
Out[11]: <matplotlib.figure.Figure at 0x7fb735f11dd0>
In [12]: sp.plot()
Logging¶
When imported, MPDAF initialize a logger by default. This logger uses the
logging module, and log messages to stderr, for instance for the .info()
methods. See Logging (mpdaf.log) for more details.
Online Help¶
Because different sub-packages have very different functionality,
further suggestions for getting started are provided in the online
documentation of these sub-packages. For example, click on Cube object,
Image object, or Spectrum object for help with the 3 main classes of
the mpdaf.obj package.
Alternatively, if you use the IPython interactive python shell, then you can
look at the docstrings of classes, objects and functions by following them with
the magic ? of IPython. Examples of this are shown below. A more general
way to see these docstrings, which works in all Python shells, is to use the
built-in help() function:
In [13]: Cube.sum?
Signature: Cube.sum(self, axis=None, weights=None)
Docstring:
Return a sum over the given axis.
Parameters
----------
axis : None or int or tuple of ints
Axis or axes along which a sum is performed:
- The default (axis = None) performs a sum over all the
dimensions of the cube and returns a float.
- axis = 0 performs a sum over the wavelength dimension and
returns an image.
- axis = (1,2) performs a sum over the (X,Y) axes and returns
a spectrum.
Other cases return None.
weights : ndarray, np.ma.array, float
When an array of weights is provided via this argument, it
used to perform a weighted sum. This involves obtaining a
weighted mean using the Cube.mean() function, then scaling
this by the number of points that were averaged along the
specified axes. The number of points that is used to scale
the mean to a sum, is the total number of points along the
averaged axes, not the number of unmasked points that had
finite weights. As a result, the sum behaves as though all
pixels along the averaged axes had values equal to the
mean, regardless of whether any of these were masked or
had zero weight.
The weights array can have the same shape as the cube, or
they can be 1-D if axis=(1,2), or 2-D if axis=0. If
weights=None, then all non-masked data points are given a
weight equal to one. Finally, if a scalar float is given,
then the data are all weighted equally. This can be used
to get an unweighted sum that behaves as though masked
pixels in the input cube had been filled with the mean
along the averaged axes before the sum was performed.
If the Cube provides an array of variances for each
data-point, then a good choice for the array of weights is
the reciprocal of this array, (ie. weights=1.0/cube.var).
However note that not all data-sets provide variance
information, so check that cube.var is not None before
trying this.
Any weight elements that are masked, infinite or nan, are
replaced with zero. As a result, if the weights are
specified as 1.0/cube.var, then any zero-valued variances
will not produce infinite weights.
File: ~/checkouts/readthedocs.org/user_builds/mpdaf/conda/2.0/lib/python2.7/site-packages/mpdaf-2.0-py2.7-linux-x86_64.egg/mpdaf/obj/cube.py
Type: instancemethod
In [14]: help(ima.plot)
Help on method plot in module mpdaf.obj.image:
plot(self, title=None, scale='linear', vmin=None, vmax=None, zscale=False, colorbar=None, var=False, show_xlabel=True, show_ylabel=True, ax=None, unit=Unit("deg"), **kwargs) method of mpdaf.obj.image.Image instance
Plot the image with axes labeled in pixels. If either axis
has just one pixel, plot a line instead of an image.
Colors are assigned to each pixel value as follows. First each
pixel value, pv, is normalized over the range vmin to vmax,
to have a value nv, that goes from 0 to 1, as follows:
nv = (pv - vmin) / (vmax - vmin)
This value is then mapped to another number between 0 and 1
which determines a position along the colorbar, and thus the
color to give the displayed pixel. The mapping from normalized
values to colorbar position, color, can be chosen using the
scale argument, from the following options:
'linear' => color = nv
'log' => color = log(1000 * nv + 1) / log(1000 + 1)
'sqrt' => color = sqrt(nv)
'arcsinh' => color = arcsinh(10*nv) / arcsinh(10.0)
A colorbar can optionally be drawn. If the colorbar
argument is given the value 'h', then a colorbar is drawn
horizontally, above the plot. If it is 'v', the colorbar
is drawn vertically, to the right of the plot.
By default the image image is displayed in its own
plot. Alternatively to make it a subplot of a larger figure, a
suitable matplotlib.axes.Axes object can be passed via the ax
argument. Note that unless matplotlib interative mode has
previously been enabled by calling matplotlib.pyplot.ion(),
the plot window will not appear until the next time that
matplotlib.pyplot.show() is called. So to arrange that a new
window appears as soon as Image.plot() is called, do the
following before the first call to Image.plot().
import matplotlib.pyplot as plt
plt.ion()
Parameters
----------
title : str
An optional title for the figure (None by default).
scale : 'linear' | 'log' | 'sqrt' | 'arcsinh'
The stretch function to use mapping pixel values to
colors (The default is 'linear'). The pixel values are
first normalized to range from 0 for values <= vmin,
to 1 for values >= vmax, then the stretch algorithm maps
these normalized values, nv, to a position p from 0 to 1
along the colorbar, as follows:
linear: p = nv
log: p = log(1000 * nv + 1) / log(1000 + 1)
sqrt: p = sqrt(nv)
arcsinh: p = arcsinh(10*nv) / arcsinh(10.0)
vmin : float
Pixels that have values <= vmin are given the color
at the dark end of the color bar. Pixel values between
vmin and vmax are given colors along the colorbar according
to the mapping algorithm specified by the scale argument.
vmax : float
Pixels that have values >= vmax are given the color
at the bright end of the color bar. If None, vmax is
set to the maximum pixel value in the image.
zscale : bool
If True, vmin and vmax are automatically computed
using the IRAF zscale algorithm.
colorbar : str
If 'h', a horizontal colorbar is drawn above the image.
If 'v', a vertical colorbar is drawn to the right of the image.
If None (the default), no colorbar is drawn.
ax : matplotlib.Axes
An optional Axes instance in which to draw the image,
or None to have one created using matplotlib.pyplot.gca().
unit : `astropy.units.Unit`
The units to use for displaying world coordinates
(degrees by default). In the interactive plot, when
the mouse pointer is over a pixel in the image the
coordinates of the pixel are shown using these units,
along with the pixel value.
kwargs : matplotlib.artist.Artist
Optional extra keyword/value arguments to be passed to
the ax.imshow() function.
Returns
-------
out : matplotlib AxesImage